验证
由于文章没上传验证数据集数据,用 p = 0.7参数从clin_mRNA_miRNA中 随机抽取取70%做训练集,其余为验证集
if(!require(caret))(install.packages(caret))
set.seed(121)
samdata<- createDataPartition(clin_mRNA_miRNA$`EVENT`, p=0.7, list=F)
valid<-clin_mRNA_miRNA[-samdata,]
summary(valid)
#单因素分析
sur2<-Surv(time=valid$`OS`,event = valid$`EVENT`)#数据集需要修改
#批量多个单因素cox回归
univarcox<- function(x){
formu<-as.formula(paste0('sur2~',x))
unicox<-coxph(formu,data=valid)##数据集更改
unisum<-summary(unicox)#汇总数据
HR<-round(unisum$coefficients[,2],3)# HR风险比
Pvalue<-unisum$coefficients[,5]#p值
CI95<-paste0(round(unisum$conf.int[,c(3,4)],3),collapse = '-') #95%置信区间
univarcox<-data.frame('characteristics'=x,
'Hazard Ration'= HR,
'CI95'= CI95,
'pvalue'= ifelse(Pvalue < 0.001, "< 0.001", round(Pvalue,3)))
return(univarcox)#返回数据表
}
#选择需要进行单因素分析变量名称
(variable_names<-colnames(valid)[c(2:9,12,13)])
univar<-lapply(variable_names,univarcox)
univartable<-do.call(rbind,lapply(univar,data.frame))
univartable$`HR(95%CI)`<-paste0(univartable$Hazard.Ration,'(',univartable$CI95,')')
univartable2<-dplyr::select(univartable,characteristics,`HR(95%CI)`,pvalue,-CI95,-Hazard.Ration)
#多因素分析
#选取p值<0.05的因素
(names2<-as.character(univartable2$characteristics[univartable2$pvalue<0.05]))
##多因素cox回归
#整合多个因素 p<0.05的公式
(form<-as.formula(paste0('sur2~',paste0(names2,collapse = '+'))))
multicox<-coxph(formula = form,data = valid)#数据集需要更改
multisum<-summary(multicox)##汇总
muHR<-round(multisum$coefficients[,2],3)#风险比
muPvalue<-multisum$coefficients[,5]#p值
muCIdown<-round(multisum$conf.int[,3],3)#下
muCIup<-round(multisum$conf.int[,4],3)#上
muCI<-paste0(muCIdown,'-',muCIup)##95%置信区间
multicox2<-data.frame('characteristics'=names2,
'muHazard Ration'=muHR,
'muCI95'=muCI,
'mupvalue'=ifelse(muPvalue < 0.001, "< 0.001", round(muPvalue,3)))
rownames(multicox2)<-NULL
multicox2$`HR(95%CI)`<-paste0(multicox2$muHazard.Ration,'(',multicox2$muCI95,')')
multicox2<-dplyr::select(multicox2,characteristics,`HR(95%CI)`,mupvalue,-muCI95,-muHazard.Ration)
#合并验证集单因素多因素结果
uni_multi2<-dplyr::full_join(univartable2,multicox2,by="characteristics")
uni_multi2$characteristics <- as.character(uni_multi2$characteristics)
输出html格式表格
#合并TCGA与验证集单因素多因素表格
comtable<-rbind(uni_multi,uni_multi2,stringsAsFactors = F)
colnames(comtable)<-c("","Univariate analysis\nHR (95% CI)","\nP value","Multivariate analysis\nHR (95% CI)","\nP value")
#表格里面不打印NA
comtable[is.na(comtable)] <- ""
#生成html
require(kableExtra)
# if (knitr:::is_html_output()) {
# cn = sub("\n", "<br>", colnames(comtable))
# } else if (knitr:::is_latex_output()) {
# usepackage_latex('makecell')
# usepackage_latex('booktabs')
# cn = linebreak(colnames(comtable), align="c")
# }
cn = sub("\n", "<br>", colnames(comtable))
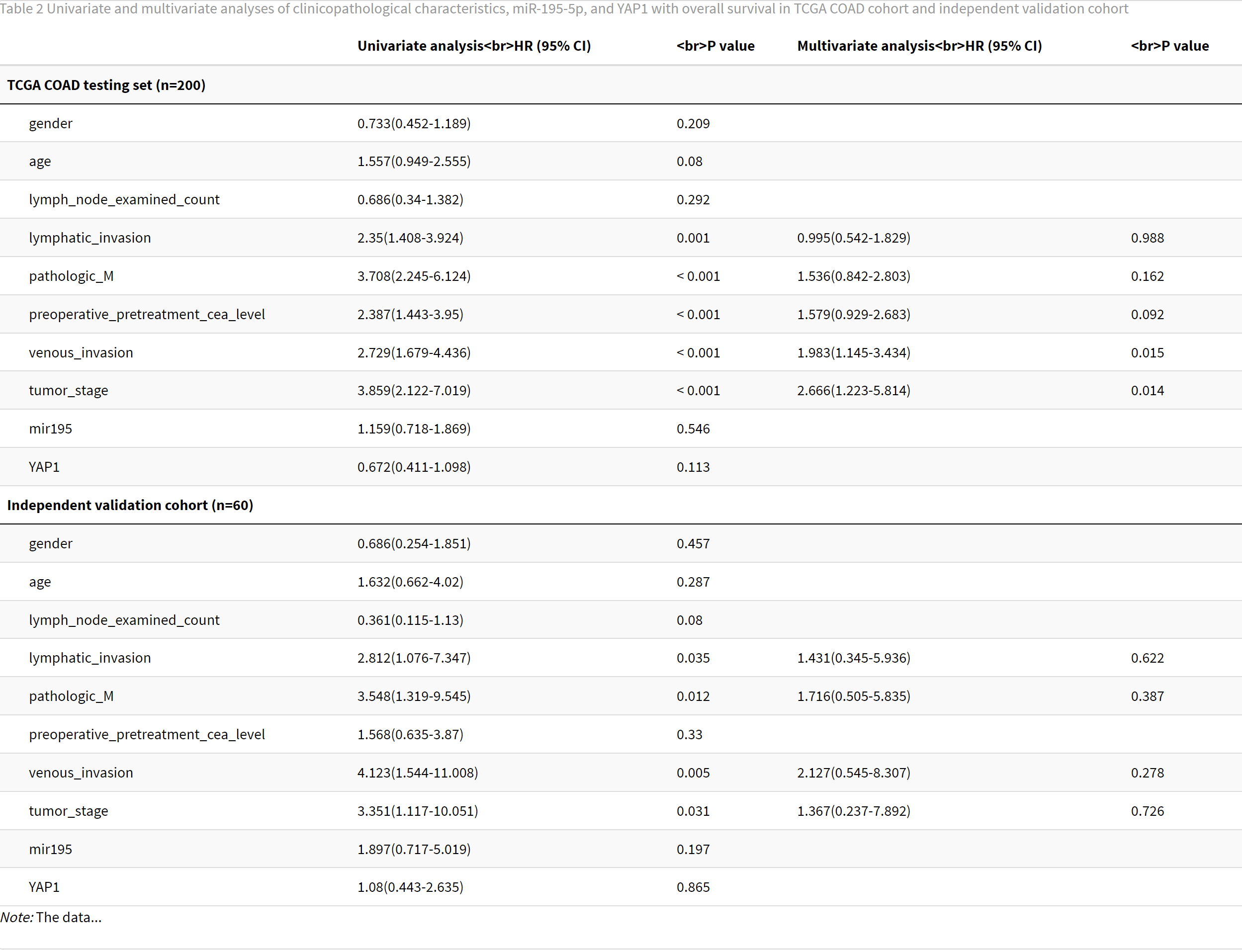
comtable %>%
kable(booktabs = T, escape = F, caption = "Table 2 Univariate and multivariate analyses of clinicopathological
characteristics, miR-195-5p, and YAP1 with overall survival in TCGA COAD
cohort and independent validation cohort",
col.names = cn) %>%
kable_styling(c("striped", "scale_down")) %>%
group_rows("TCGA COAD testing set (n=200)", 1,nrow(uni_multi)) %>%
group_rows("Independent validation cohort (n=60)", nrow(uni_multi)+1,nrow(comtable)) %>%
footnote(general = "The data...",
footnote_as_chunk = T)
生成csv格式的表格
#合并TCGA与验证集单因素多因素表格
table_subtitle <- c(NA,"HR (95% CI)","P value","HR (95% CI)","P value")
TCGA <- c("TCGA COAD testing set (n=200)",rep("",4))
val<-c("Independent validation cohort (n = 60)",rep("",4))
comtable<-rbind(table_subtitle,TCGA, uni_multi,val,uni_multi2,stringsAsFactors = F)
colnames(comtable)<-c("","Univariate analysis","","Multivariate analysis","")
#表格里面不打印NA
comtable[is.na(comtable)] <- ""
str(comtable)
#保存到csv文件
write.csv(comtable,"Table2.csv", quote = F, row.names = F)
生成word格式的表格(推荐,投稿可能有用)
#合并TCGA与验证集单因素多因素表格
table_subtitle <- c(NA,"HR (95% CI)","P value","HR (95% CI)","P value")
TCGA <- c("TCGA COAD testing set (n=200)",rep(NA,4))
val<-c("Independent validation cohort (n = 60)",rep(NA,4))
comtable<-rbind(table_subtitle,TCGA, uni_multi,val,uni_multi2,stringsAsFactors = F)
colnames(comtable)<-c(NA,"Univariate analysis",NA,"Multivariate analysis",NA)
#表格里面不打印NA
comtable[is.na(comtable)] <- ""
#保存到word文档
title_name<-'Table 2 Univariate and multivariate analyses of clinicopathological
characteristics, miR-195-5p, and YAP1 with overall survival in TCGA COAD
cohort and independent validation cohort'
table1<-comtable
mynote <- "Note: ..."
if(!require(officer)) (install.packages('officer'))
library(officer)
my_doc <- read_docx() #初始化一个docx
my_doc %>%
##添加段落标题名称
body_add_par(value = title_name, style = "table title") %>%
#添加表格
body_add_table(value = table1, style = "Light List Accent 2" ) %>%
#添加Note
body_add_par(value = mynote) %>%
#打印到word文档
print(target = "Table2.docx")
#查看表格相关参数
read_docx() %>% styles_info() %>%
subset( style_type %in% "table" )
#用这些参数,把表格设置成你想要的形式
 |Archiver|手机版|小黑屋|bioinfoer
( 萌ICP备20244422号 )
|Archiver|手机版|小黑屋|bioinfoer
( 萌ICP备20244422号 )