背景
图一:不同的癌症用不同的颜色展示
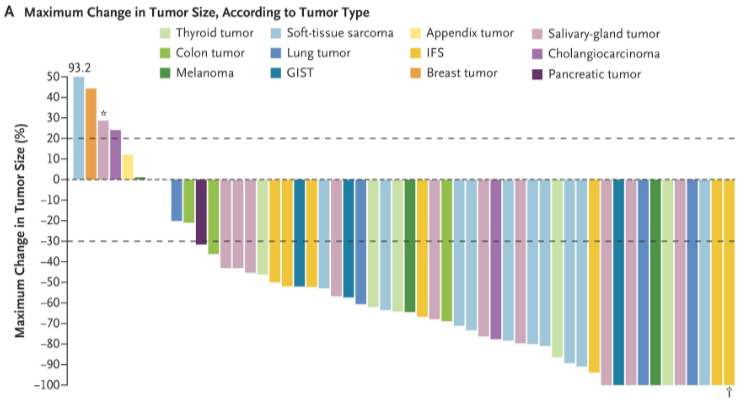
出自文章PMID: 29466156
图二:做类似的图用于GO、KEGG富集分析结果展示,不显著的显示灰色,左右分开的名称,正负富集分开,左右分开画在一张图上。
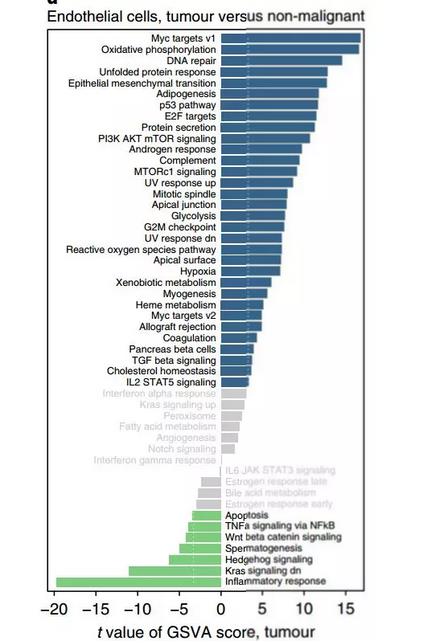
出自文章PMID: 29988129
原文的输入数据是GSVA score,本代码只模仿形式,不包括GSVA分析。
应用场景
场景一:各肿瘤某一指标的比较,例如图一的肿瘤缓解率。
场景二:GSVA结果展示,类似于图二。
场景一
每个ID显示为一种颜色
输入数据
df<-read.csv("easy_input1.csv")
head(df)
#按照score排序
df<-df[order(df$score,decreasing = T),]
df$index<-seq(1,nrow(df))
head(df)
开始画图
library(ggplot2)
p<-ggplot(df,aes(x=index,y=score,fill=ID)) +
geom_bar(stat = 'identity',width = 0.8) +
scale_fill_brewer(type = "Qualitative", palette = "Paired") + #bar的颜色
scale_y_continuous(breaks=seq(-100, 100, 10), #y轴刻度
expand = c(0,0)) + #上下都不留空
scale_x_discrete(expand = expand_scale(mult = c(0.01,0))) + #左边留空,右边到头
#画3条横线
geom_hline(yintercept = c(-30,0,20),
linetype = 5, #画虚线
size = 0.3) + #线的粗细
#其他主题
labs(x = "", y = "Maximum Change in Tumor Size (%)",
title = "A Maximum Change in Tumor Size, According to Tumor Type") +
theme_bw() + #去除背景色
theme(panel.grid =element_blank()) + #去除网格线
theme(panel.border = element_blank()) + #去除外层边框
theme(axis.line = element_line(colour = "black")) + #沿坐标轴显示直线
theme(axis.line.x = element_blank(), axis.ticks.x = element_blank(), axis.text.x = element_blank()) + #去除x轴
#图例
guides(fill = guide_legend(ncol = 5,title = NULL)) + #图例分5列
scale_size(range=c(5,20)) +
theme(legend.background = element_blank(), #移除整体边框
#图例的左下角置于绘图区域的左下角
legend.position=c(0,0),legend.justification = c(0,0))
#改用下面这行,图例就会位于顶部
#legend.position="top")
p
#保存到PDF文件
ggsave("CancerBar.pdf",width = 8,height = 5)
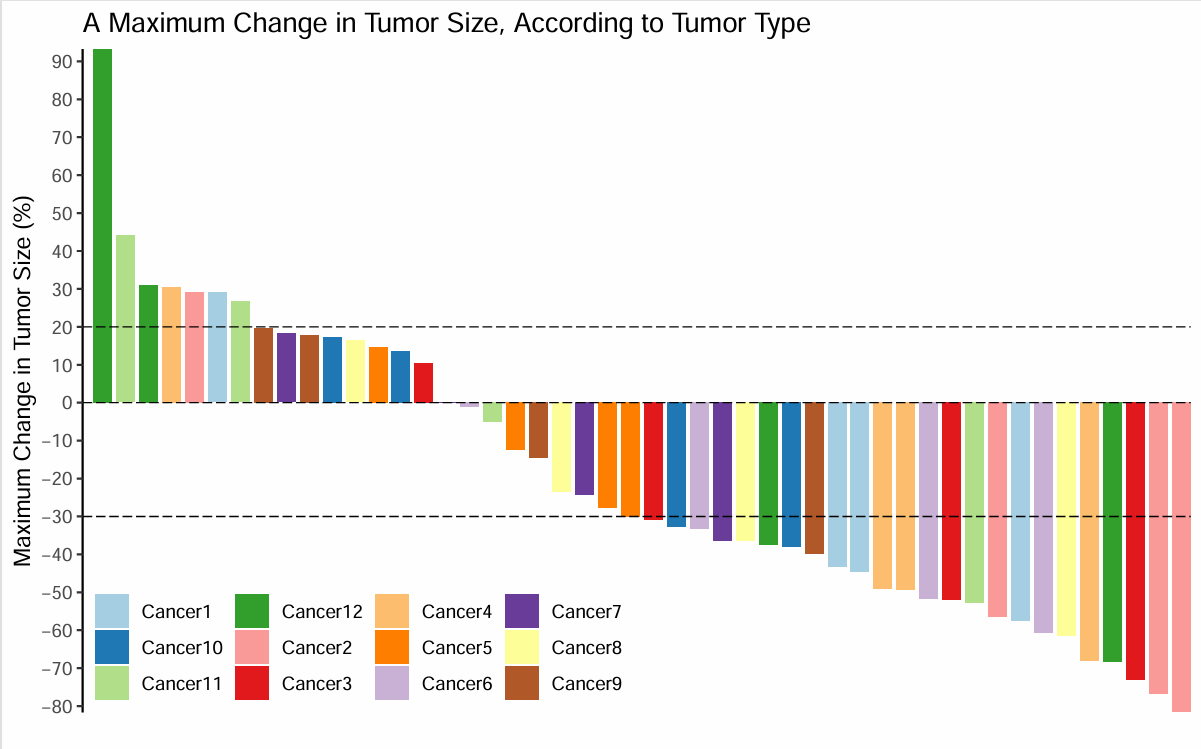
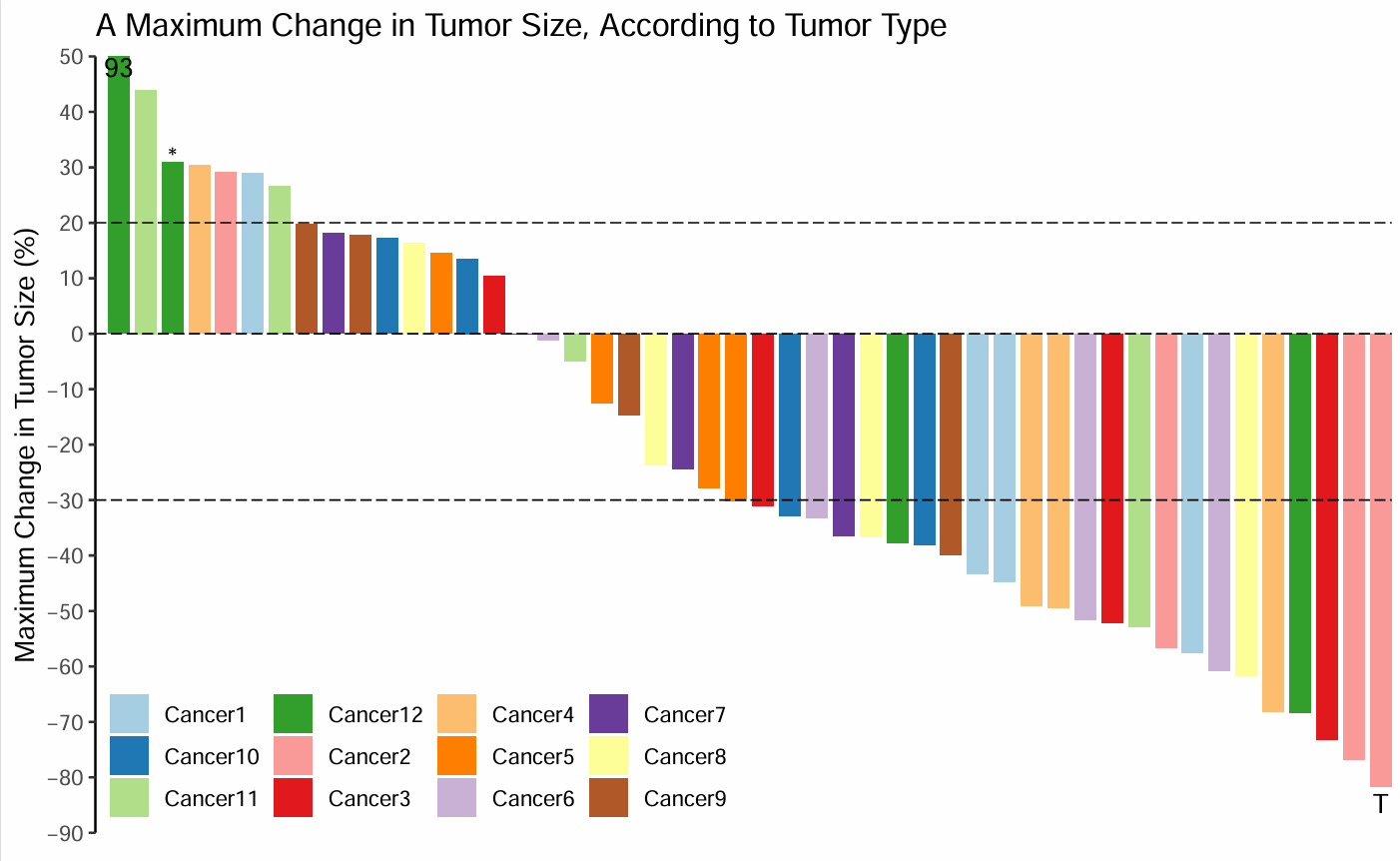
极大值的处理
有一个值远高于其他值,右侧会很空。
因此,原图作者让y轴适合大部分数据,然后在最高的那个bar上标出实际数据
#设置坐标轴范围,最大值设为50,以适应大多数数据
p<-p + coord_cartesian(ylim = c(-90,50)) + #y轴范围,根据实际情况调整
#添加数据标签
geom_text(data = subset(df, score > 50),
aes(index, 48,label=round(score))) + #在超过50的bar上标出实际数据
geom_text(data = subset(df, index == 3),
aes(index, score + 1,label = "*")) + #作者的特殊标记
geom_text(data = subset(df, index == nrow(df)),
aes(index, score - 3, label = "T")) #作者的特殊标记
p
#保存到PDF文件
ggsave("CancerBar_label.pdf",width = 8,height = 5)
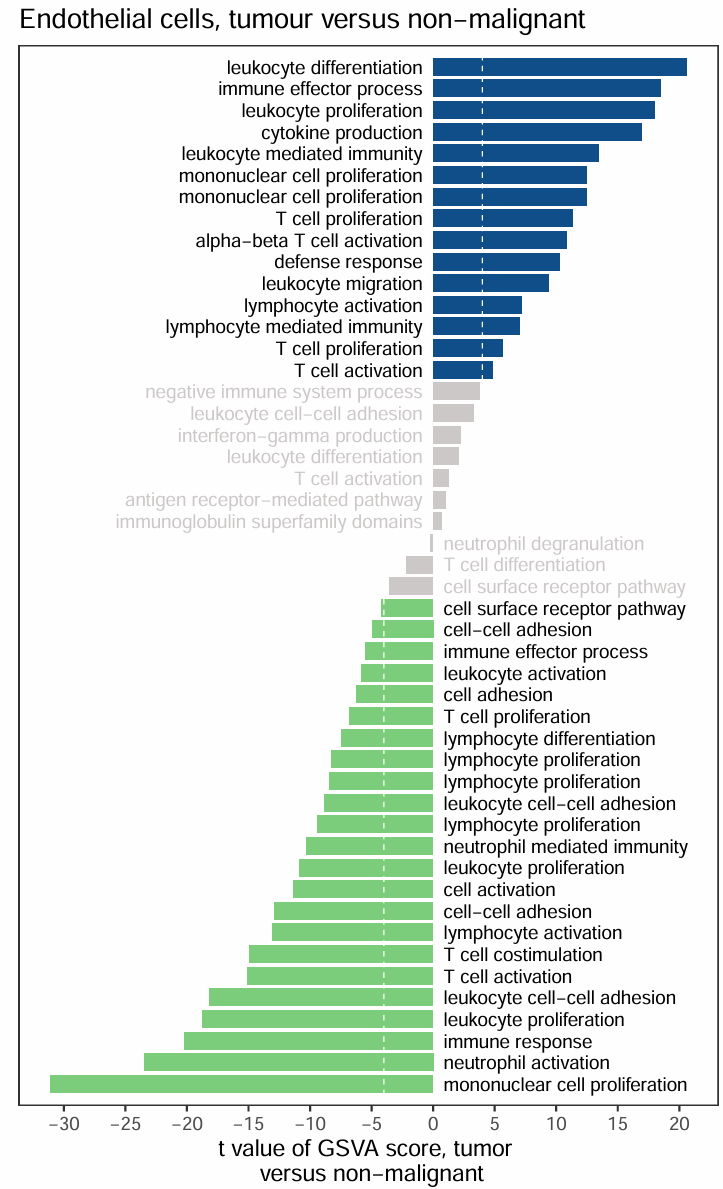
场景二·原文
score绝对值小于阈值的bar显示为灰色
输入数据
包含两列:ID和score
df<-read.csv("easy_input2.csv")
head(df)
#按照score的值分组
df$group<-cut(df$score, breaks = c(-Inf,-4,4,Inf),labels = c(1,2,3))
head(df)
#按照score排序
df<-df[order(df$score,decreasing = F),]
df$index<-seq(1,nrow(df))
head(df)
开始画图
library(ggplot2)
ggplot(df,aes(x=index,y=score,fill=group)) +
geom_bar(stat = 'identity',width = 0.8) +
scale_fill_manual(values = c("palegreen3","snow3","dodgerblue4")) + #bar的颜色
scale_x_discrete(expand = expand_scale(add = .6)) +
scale_y_continuous(breaks=seq(-30, 20, 5)) +
coord_flip() + #坐标轴互换
#画2条横线
geom_hline(yintercept = c(-4,4),
color="white",
linetype = 2,#画虚线
size = 0.3) + #线的粗细
#写label
geom_text(data = subset(df, score > 0),
aes(x=index, y=0, label=paste0(ID," "), color = group),#bar跟坐标轴间留出间隙
size = 3, #字的大小
hjust = "inward" ) + #字的对齐方式
geom_text(data = subset(df, score < 0),
aes(x=index, y=0, label=paste0(" ",ID), color = group),
size = 3, hjust = "outward") +
# 注意标签的黑色灰色颜色顺序
scale_colour_manual(values = c("snow3","black","black")) +
# scale_colour_manual(values = c("black","snow3","black")) +
#其他主题
labs(x = "", y = "t value of GSVA score, tumor \n versus non-malignant",
title = "Endothelial cells, tumour versus non-malignant") +
theme_bw() + #去除背景色
theme(panel.grid =element_blank()) + #去除网格线
theme(panel.border = element_rect(size = 0.6)) + #边框粗细
theme(axis.line.y = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank()) + #去除y轴
guides(fill=FALSE,color=FALSE) #不显示图例
#保存到PDF文件
ggsave("GObar.pdf",width = 5,height = 8)
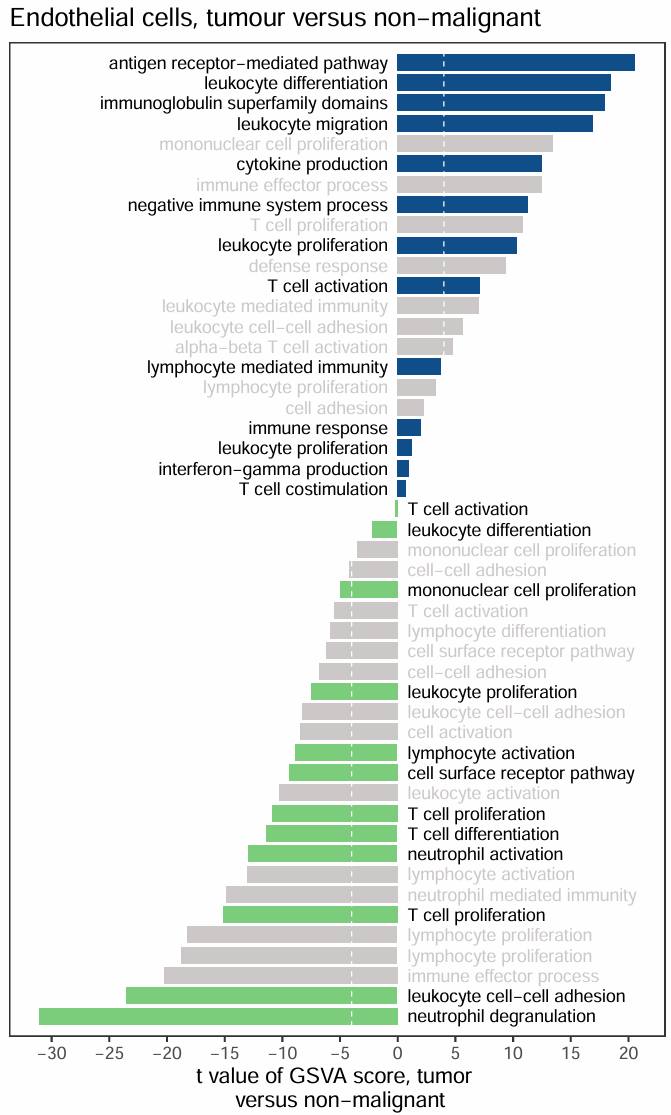
场景二·不显著的显示为灰色
pvalue>0.05的bar显示为灰色
输入数据
包含三列,ID、score和pvalue
df<-read.csv("easy_input3.csv")
head(df)
#按照pvalue分组
df$p.group<-cut(df$pval, breaks = c(-Inf,0.05,Inf),labels = c(1,0))
#按照score分组
df$s.group<-cut(df$score, breaks = c(-Inf,0,Inf),labels = c(0,1))
#合并
df$ps.group <- paste0(df$p.group,df$s.group)
#根据pvalue和score分为3组
df$group <- ifelse(df$ps.group=='10','1',ifelse(df$ps.group=='11','2','3'))
head(df)
#按照score排序
df<-df[order(df$score,decreasing = F),]
df$index<-seq(1,nrow(df))
head(df)
开始画图
只调整了颜色顺序,其余跟“场景二·原文”的画图代码是一样的
library(ggplot2)
ggplot(df,aes(x=index,y=score,fill=group)) +
geom_bar(stat = 'identity',width = 0.8) +
scale_fill_manual(values = c("palegreen3","dodgerblue4","snow3")) + #颜色
scale_x_discrete(expand = expand_scale(add = .6)) +
scale_y_continuous(breaks=seq(-30, 20, 5)) +
coord_flip() +
geom_hline(yintercept = c(-4,4),
color="white",
linetype = 2,
size = 0.3) +
geom_text(data = subset(df, score > 0),
aes(x=index, y=0, label=paste0(ID," "), color = p.group),
size=3,
hjust = "inward") +
geom_text(data = subset(df, score < 0),
aes(x=index, y=0, label=paste0(" ",ID), color = p.group),
size=3,hjust = "outward") +
scale_colour_manual(values = c("black","snow3")) +
labs(x = "", y = "t value of GSVA score, tumor \n versus non-malignant",
title = "Endothelial cells, tumour versus non-malignant") +
theme_bw() +
theme(panel.grid =element_blank()) +
theme(panel.border = element_rect(size = 0.6)) +
theme(axis.line.y = element_blank(), axis.ticks.y = element_blank(), axis.text.y = element_blank()) +
guides(fill=FALSE,color=FALSE)
#保存到PDF文件
ggsave("GObar_pval.pdf",width = 5,height = 8)
 |Archiver|手机版|小黑屋|bioinfoer
( 萌ICP备20244422号 )
|Archiver|手机版|小黑屋|bioinfoer
( 萌ICP备20244422号 )